Pathogenesis and biomarkers of colorectal cancer by epigenetic alteration
Article information
Abstract
Colorectal cancer (CRC) ranks third in cancer incidence and stands as the second leading cause of cancer-related deaths globally. CRC tumorigenesis results from a cumulative set of genetic and epigenetic alterations, disrupting cancer-regulatory processes like cell proliferation, metabolism, angiogenesis, cell death, invasion, and metastasis. Key epigenetic modifications observed in cancers encompass abnormal DNA methylation, atypical histone modifications, and irregularities in noncoding RNAs, such as microRNAs and long noncoding RNAs. The advancement in genomic technologies has positioned these genetic and epigenetic shifts as potential clinical biomarkers for CRC patients. This review concisely covers the fundamental principles of CRC-associated epigenetic changes, and examines in detail their emerging role as biomarkers for early detection, prognosis, and treatment response prediction.
INTRODUCTION
Globally, colorectal cancer (CRC) occupies the third spot for cancer incidence and is the second leading cause of cancer-related deaths [1]. Alarmingly, CRC’s incidence is surging in numerous Asian nations [2]. Despite a noticeable decline starting from 2011, CRC remained among the top 5 diagnosed cancers in Korea in 2019. Furthermore, in Korea, CRC was the primary cause of cancer-related fatalities in men, and the second in women [3].
CRC tumorigenesis is a multi-step process, propelled by progressive accumulations of genetic and epigenetic alterations, which cause a disruption of cancer-controlling mechanisms, such as cell growth, metabolism, angiogenesis, apoptosis, invasion, and metastasis. Epigenetics, which refers to inheritable changes in gene expression that do not arise from changes in the gene’s primary nucleotide sequence, play a pivotal role. Notably, in cancers, including CRC, epigenetic alterations surface early and outnumber genetic changes [4,5]. Such epigenetic shifts involve irregular DNA methylation, aberrant histone modifications, and inconsistencies in noncoding RNAs, such as microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) (Fig. 1) [6].
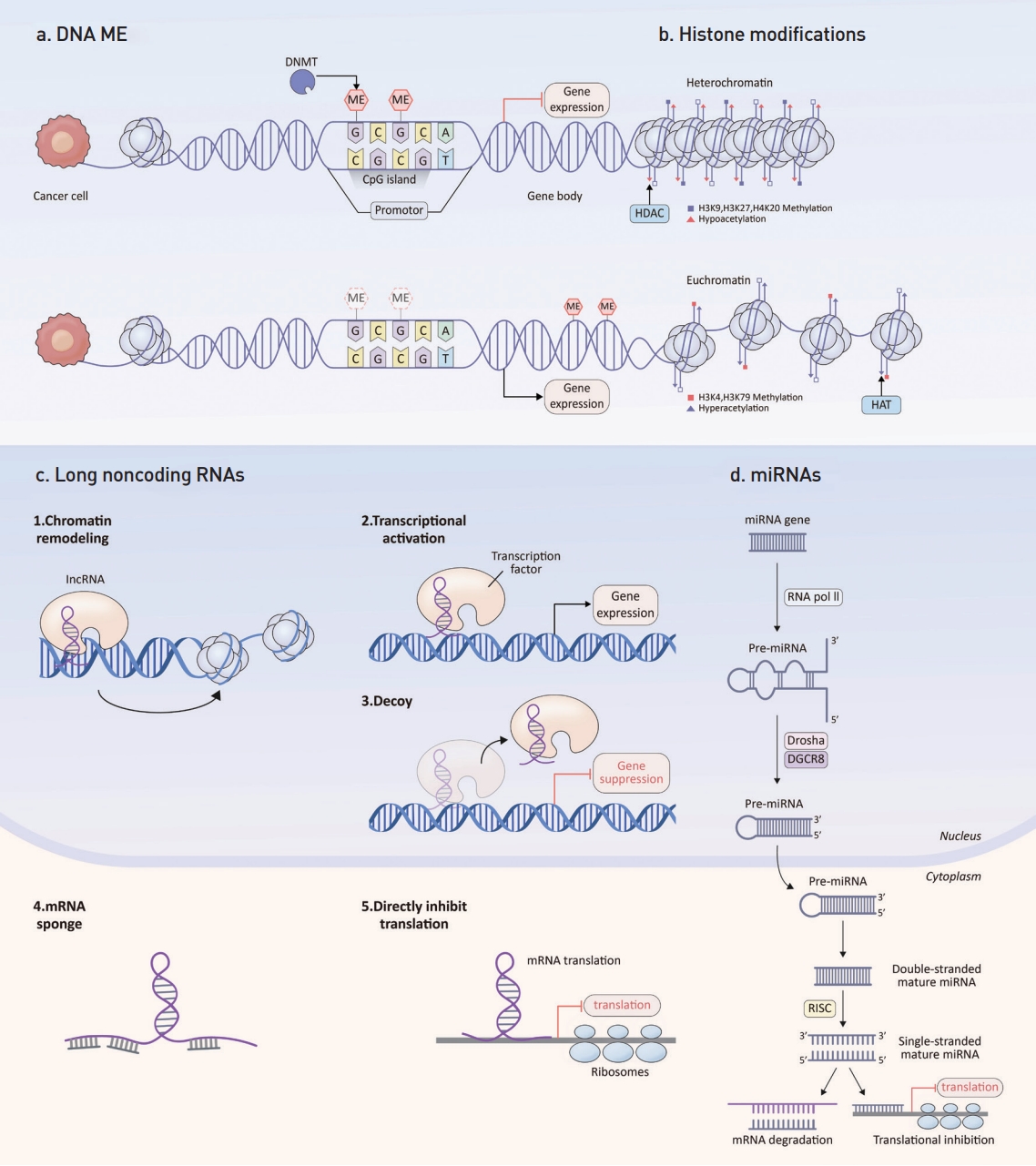
Schematic diagram of epigenetic shifts. (a) DNA ME mainly occurs in CpG islands is facilitated by DNA methyltransferases (DNMTs). This ME inhibits gene expression. (b) Histone modifications affect structure of chromatin. Heterochromatin is often associated with hypoacetylation by HDAC and inactive gene transcription. In contrast, euchromatin is associated with hyperacetylation by HAT and active gene transcription. (c) Long noncoding RNAs (lncRNA) regulate chromatin remodeling, induce transcriptional activation, function as decoys and inhibits gene transcription, act as microRNA (miRNA) sponges and, directly inhibit messenger RNA (mRNA) translation. (d) miRNAs commence with RNA polymerase II (RNA Pol II) transcribing the miRNA gene. The Drosha–DGCR8 complex process this into pre-miRNA, which is then transported to the cytoplasm by exportin 5. And then, RNase III enzyme DICER produces a double-stranded mature miRNA. The RNA-induced silencing complex (RISC) integrates one of the strands and facilitates its interaction with the target mRNA, leading to either translational inhibition or mRNA degradation. ME, methylation; HDAC, histone deacetylases; HAT, histone acetyltransferases.
Colonoscopy stands as the gold standard for CRC screening but has several drawbacks, such as the need for bowel preparation, potential side effects from sedation, associated costs, and low patient compliance. A study by the Nordic-European Initiative on Colorectal Cancer highlighted that colonoscopy could curtail CRC risk by 18% over a decade. Yet, only 42% of the invited participants opted for the screening [7]. The fecal immunohistochemical test (FIT), a global choice for noninvasive CRC screening, demands enhancements in sensitivity. Molecular screening approaches like the multi-target stool DNA test, based on methylated DNA, hold more promise than FIT. Nevertheless, the financial strain of such tests remains a barrier to their widespread adoption [8].
Modern genomic technologies have unveiled numerous genetic and epigenetic variations, positioning them as hopeful clinical indicators for CRC patients. This review aimed to elucidate the foundational aspects of CRC-related epigenetic modifications and evaluate their potential as biomarkers in the arenas of early detection, prognosis, and therapeutic prediction.
METHODS
In this review, articles detailing the use of epigenetic alterations as biomarkers for diagnosis, prognosis, and treatment prediction in CRC were examined and discussed. Comprehensive searches were conducted in medical databases, including PubMed, Embase, Scopus, and Google Scholar, using keywords such as colorectal cancer, epigenetic alteration, DNA methylation, histone modification, miRNA, lncRNA, biomarker, diagnosis, prognosis, and prediction. All pertinent articles have been incorporated into this review.
DNA METHYLATION
In eukaryotic cells, DNA methylation mainly occurs at the 5-prime position of the cytosine ring within CpG dinucleotides. This methylation modulates gene transcription by impacting both promoter regions and noncoding DNA segments, such as enhancers. The methylation of 5-cytosine is facilitated by DNA methyltransferases (DNMTs) [9]. Predominantly, DNA methylation is found within repetitive genomic areas, which encompass satellite DNA and parasitic sequences like long interspersed transposable elements (LINEs) and short interspersed transposable elements (SINEs) [10]. It can directly impede gene expression by obstructing specific transcription factor binding. It can also indirectly influence gene expression by recruiting methyl-CpG-binding domain (MBD) proteins [1]. One of the initial aberrant methylation changes in CRC is global DNA hypomethylation. LINE-1 sequence hypomethylation might correlate with genomic instabilities, including microsatellite instability (MSI) and the CpG island methylator phenotype (CIMP) [2,3]. Moreover, promoter hypermethylation is correlated with the silencing of tumor suppressor genes, inducing oncogenesis by affecting essential cellular mechanisms such as DNA repair, cell cycle control, apoptosis, angiogenesis, and tumor invasion [4,5]. Aberrant DNA methylation markers have demonstrated clinical relevance as diagnostic indicators. Additionally, these DNA methylation markers have potential as prognostic biomarkers.
1. Blood-Based Diagnostic DNA Methylation Biomarkers
DNA methylation typically manifests in the early stages of CRC and can be potential early risk indicators. Currently, the most recognized blood-based diagnostic DNA methylation biomarker is methylated Septin 9 (SEPT9), a gene encoding GTP-binding proteins linked with cytoskeletal remodeling. The diagnostic accuracy of this biomarker has been confirmed in several CRC studies, displaying a sensitivity range from 48.2% to 95.6% and specificity between 79.1% and 99% [6-16]. Commercially, it is available as Epi proColon® (Epigenomics) and received Food and Drug Administration (FDA) approval in 2016 for CRC screening. However, major limitation of methylated SEPT9 is its relatively low sensitivity in diagnosing advanced adenomas, ranging from 7.9% to 38.7% [7,17-21]. Another promising DNA methylation biomarker is methylated secreted frizzled-related protein 2 (SFRP2), which acts as a modulator of Wnt signaling. Two studies reported its diagnostic accuracy in CRC, revealing sensitivity from 63.8% to 66.9% and specificity between 97.3% and 100% [22,23]. The Syndecan 2 (SDC2) gene, encoding an integral membrane protein involved in cell proliferation, migration, and cell-matrix interaction, has been recognized as another potential biomarker. Studies on methylated SDC2 reported a sensitivity between 87% to 87.2% and specificity from 95.2% to 100% for CRC [23,24].
Given the multifaceted origins of CRC, arising from an interplay of genetic mutations and epigenetic alterations, numerous studies have explored multiple epigenetic biomarkers, combined to enhance detection accuracy for advanced adenomas and CRC [23,25-28]. ColoDefense®, which combines methylated SEPT9 and SDC2 in one assay, has been introduced to improve CRC screening. Its diagnostic accuracy in CRC patients ranges in sensitivity from 86.5% to 88.9% and specificity from 92.1% to 92.8%.29,30 Another combined test for CRC screening is the ColveraTM (by Clinical Genomics), which identifies methylated branched chain amino acid transaminase 1 (BCAT1) and IKAROS family zinc finger 1 (IKZF1). Pedersen et al. [31] reported its diagnostic accuracy for CRC, with sensitivity between 56% and 79% and specificity from 94% to 95%. Barták et al. [23] presented a 4-biomarker panel (SFRP1, SFRP2, SDC2, and proline rich membrane anchor 1 [PRIMA1]) with a reported sensitivity of 91.5% and specificity of 97.3% in CRC diagnosis. TriMeth, a diagnostic DNA methylation biomarker for earlystage CRC detection, utilizes a combination of 3 methylated biomarkers (chromosome 9 open reading frame 50 [C9orf50], potassium voltage-gated channel subfamily Q member 5 [KCNQ5], and CAP-Gly domain containing linker protein family member 4 [CLIP4]). Jensen et al. [32] showcased its efficacy with an overall sensitivity of 85% (stage I: 80%; stage II: 85%; stage III: 89%; stage IV: 88%) and a specificity of 99% in an independent cohort. Multiple blood-based diagnostic DNA methylation biomarkers, including aristaless-like homeobox 4 (ALX4), adenomatous polyposis coli (APC), cyclin dependent kinase inhibitor 2A (CDKN2A), helicase-like transcription factor (HLTF), hyperpigmentation, progressive, 1 (HPP1), MutL homolog-1 (MLH1), methylguanine methyltransferase (MGMT), N-Myc, downstream-regulated gene-4 (NDRG4), Neurogenin-1 (NEUROG1), and nerve growth factor receptor (NGFR), Ras association domain family member-2, isoform-A (RASSF2A), transmembrane protein with epidermal growth factor (EGF) like and two follistatin like domains 2 (TMEFF2), vimentin (VIM), and Wnt inhibitory factor 1 (WIF1), have been explored for CRC screening [20,25,26,33-41]. Table 1 summarizes the most promising DNA methylation genes and panels as potential blood-based diagnostic biomarkers for CRC.
2. Prognostic and Predictive DNA Methylation Biomarkers
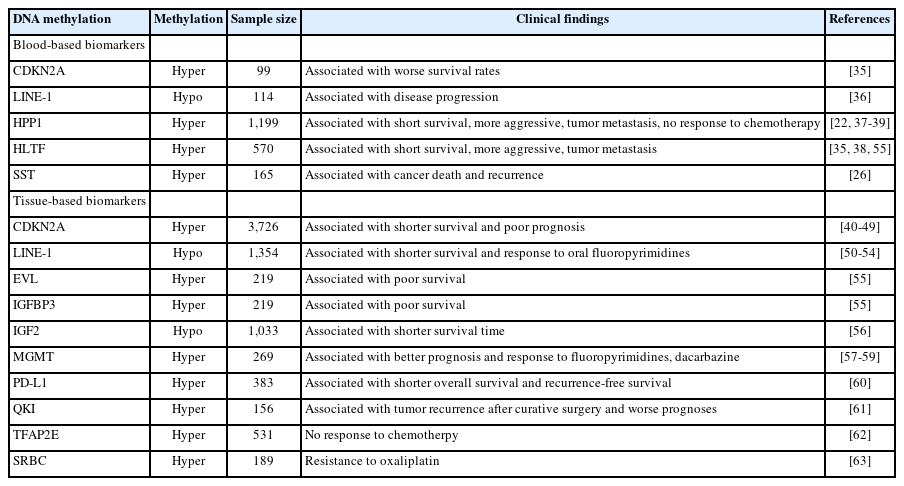
CDKN2A hypermethylation is among the extensively studied biomarkers. It is linked with poor prognosis, an elevated risk of recurrence, and metastasis in CRC patients [34,42-49]. Similarly, LINE-1 hypomethylation, another well-studied marker, correlates with adverse outcomes in CRC patients [50-53]. Moreover, LINE-1 has demonstrated a survival advantage in CRC patients who have undergone oral fluoropyrimidines treatment [54]. Recent studies have highlighted HPP1 and HLTF as prognostic biomarkers for CRC. Hypermethylation of HPP1 and HLTF correlates with advanced CRC (stages III and IV), adverse outcomes, and recurrence [35,55-57]. Yi et al. [58] found DNA methylation of insulin-like growth factor binding protein 3 (IGFBP3) and Enah/Vasp-like (EVL) associated with negative outcomes. In their study, DNA hypermethylation of certain extracellular matrix (ECM) genes was notably linked with diminished survival. Hypermethylation of MGMT, which is instrumental in defending cells from mutagenesis and alkylating agents, aligns with a positive prognosis in CRC patients treated with 5-fluorouracil (5-FU) and dacarbazine [59,60]. These studies provide evidence that aberrantly methylated DNA has the potential to be used as prognostic and predictive biomarkers for CRC. Further large-scale clinical studies for validation are essential. Other potential prognostic and predictive DNA methylation biomarkers under study include insulin-like growth factor 2 (IGF2), programmed cell death-ligand 1 (PD-L1), RNA-binding protein quaking (QKI), somatostatin (SST), transcription factor AP-2 epsilon (TFAP2E), and serum deprivation response factor-related gene product that binds to the c-kinase (SRBC) [61-67]. Table 2 summarizes most promising DNA methylation genes as potential prognostic and predictive biomarkers for CRC.
3. Stool-Based Diagnostic DNA Methylation Biomarkers
Methylated VIM, encoding the intermediate filament protein vimentin that forms the cytoskeleton, is a recurrently observed biomarker for CRC diagnosis. Several studies confirm its diagnostic precision, with a sensitivity spanning from 38.3% to 81% and specificity between 82% and 95% [40,68-70]. It was the first stool-based DNA methylation biomarker approved for the early CRC detection and was commercialized as the ColoSureTM (LabCorp) [71]. DNA methylation biomarkers like NDRG4 are promising for early CRC screening. Melotte et al. reported NDRG4 methylation’s positive correlation with CRC, showcasing sensitivity of 53% and specificity of 100% in stool samples [72].
Cologuard® (Exact Sciences) is an FDA-approved multi-target stool DNA test for CRC screening. This test combines Kirsten rat sarcoma viral oncogene homologue (KRAS) mutations and bone morphogenetic protein-3 (BMP3) and NDRG4 methylation levels and including an immunochemical assay for hemoglobin. Its diagnostic efficacy for CRC is marked by sensitivity of 92.3% and specificity of 86.6%, respectively. Notably, a study reported that Cologuard was superior sensitive than FIT (42.4% vs. 23.8%) in detecting advanced adenoma or sessile serrated lesions over ≥ 1 cm [73]. Tissue factor pathway inhibitor-2 (TFPI2), a serine proteinase inhibitor, is perceived as a tumor suppressor, obstructing the degradation of cancer cells’ extracellular matrix and preventing tumor invasion. Zhang et al. presented a 2-biomarker panel (SDC2, TFPI2) for CRC, achieving sensitivity of 93.4% and specificity of 94.3% [74]. In other study, Park et al. reported a 4-biomarker panel (SFRP2, TFPI2, NDRG4, BMP3) for Korean CRC patients, with sensitivity of 94.3% and specificity of 55.0% [75]. Another study identified a stool DNA methylation panel (SFRP2, GATA binding protein 4/5 [GATA4/5], NDRG4, VIM) in CRC patients, showing sensitivity of 96.4% and specificity of 65.0% [76]. Additional stool-based diagnostic DNA methylation biomarkers under study for CRC screening encompass CDKN2A, GATA4, MLH1, integrin subunit alpha 4 (ITGA4), MGMT, oncostatin M receptor (OSMR), phosphatase and actin regulator 3 (PHACTR3), RASSF2, SEPT9, and WIF1 [19,40,68,77-87]. Table 3 summarizes most promising DNA methylation genes and panels as potential stoolbased diagnostic biomarkers for CRC.
HISTONE MODIFICATIONS
DNA is coiled around histones to form structural units termed nucleosomes. Histones are protein octamers made up of pairs of the 4 core histone proteins: histone 2A (H2A), H2B, H3, and H4. These nucleosomes, combined with other nuclear proteins, constitute chromatin. Alterations to histones influence chromatin’s structure, significantly affecting gene regulation and carcinogenesis [88,89]. The most widely studied histone modifications in CRC are histone acetylation and methylation. Enzymes known as histone acetyltransferases (HATs) and histone deacetylases (HDACs) drive histone acetylation and deacetylation, respectively. Histone acetylation impacts chromatin’s compactness. An increase in histone acetylation, associated with proto-oncogenes, prompts gene expression. In contrast, a decrease in acetylation, commonly seen in the promoter regions of tumor suppressor genes, leads to their repression, underscoring the crucial role of histone acetylation in cancer onset and progression [10]. Similarly, histone methylation affects DNA compactness and can create binding sites in the chromatin recognizable by various proteins, including transcriptional complexes. The enzymes histone methyltransferases (HMTs) and histone demethylases (HDMs) regulate histone methylation and demethylation, respectively [90]. Overexpression or underexpression of these enzymes can disturb the overall histone methylation equilibrium, altering the expression patterns of many oncogenes or tumor suppressor genes, thus influencing cancer development or progression [13]. Given their impact, histone modifications present promising opportunities as diagnostic and prognostic biomarkers in CRC.
Histone Modification Alteration as Potential Biomarkers
While there are major challenges in employing histone modifications as biomarkers, mainly due to technical restrictions such as their use as quantitative indicators and their specificity across various cancer types, considerable research has showcased their potential as CRC biomarkers. Several studies have highlighted that methylation of H3K9 and acetylation of H3K27, H4K12, H3K18 are more pronounced in CRC than in normal colonic mucosa [91-93]. Such findings hint at the potential of histone modifications as diagnostic CRC biomarkers. Additionally, methylation of H3K9, H3K27, H4K20 has been found to be notably decreased in CRC, in comparison with healthy control (HC) circulating nucleosomes [94,95]. These studies emphasize the prospective value of histone modifications as diagnostic biomarkers for CRC. Histone modifications have also been explored in the context of CRC progression and patient survival. Low methylation levels of H3K4 and H3K27 have been linked to worse survival rates [96,97]. Conversely, high methylation levels of H3K9, H3K20, and H3K27 are associated with a favorable prognosis [98-100], Several studies have identified prognostic factors via combinations of histone modifications. For instance, Benard et al. [98] found that trimethylation at H3K4, H3K9, and H4K20 relates to disease-free survival and recurrence-free survival in early-stage CRC. In their study, they revealed that a combination of histone modifications provides better patient stratification than individual markers. In another study, the expression of polycomb-group (PcG) proteins, specifically enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), B lymphoma Mo-MLV insertion region 1 polycomb ring finger (BMI1), and suppressor of zeste 12 (SUZ12), and the associated histone modification H3K27me3, were reported in CRC [101]. These were associated with the disease-free survival and the recurrence-free survival. However, these findings are preliminary and necessitate further investigations to ascertain the viability of this innovative approach. Other histone modification biomarkers studied in a CRC context include H3K56 and H4K16 [102].
MICRORNAS
MicroRNAs (miRNAs) are short (~22 nucleotides in length), single-stranded RNAs that participate in numerous cellular activities, such as development, proliferation, differentiation, apoptosis, DNA repair, and stress responses [103]. miRNA dysregulation has been linked with diverse human cancers, including CRC. miRNAs can act as tumor suppressors or oncogenes, sometimes labeled as “oncomirs.” [104] The inaugural study examining the irregular expression of miRNAs in colorectal tumor tissues demonstrated that miR-143 and miR-145 levels were substantially decreased in both precancerous adenomatous and CRC tissues relative to normal tissues. This hinted at potential alterations in the miRNA pathway during colorectal tumorigenesis [105]. A review that consolidated findings from 20 studies examining miRNA expression levels in CRC tissues found 164 dysregulated miRNAs [106]. In more than one study, miR-20a and miR-31 were found to be expressed at significantly higher levels, whereas miR-143 and miR-145 were noted to be significantly lower in CRC tissues. Collectively, miRNAs demonstrate potential as noninvasive biomarkers for early CRC diagnosis, prognosis, and predictive treatment responses.
1. Blood-Based Diagnostic miRNA Biomarkers
miRNA levels in serum, plasma, and tissue samples are notably stable. One study illustrated that extracellular miRNA remains unchanged for a minimum of 1 month, pointing to the potential use of extracellular miRNA as a cancer diagnostic biomarker [107]. Various investigations have conducted on the use of miRNA as potential noninvasive single miRNA biomarkers for CRC. Notably, miR-21 and miR-92a have been extensively studied for CRC diagnosis [1]. Elevated levels of miR-21 in CRC and adenomatous tissue position miR-21 as a promising early diagnostic biomarker in the adenoma-carcinoma progression [108]. Toiyama et al. [109] found significantly increased miR21 levels in the preoperative serum of adenoma and CRC patients. Interestingly, postoperative serum levels of miR-21 substantially decreased after curative resection. Elevated serum and tissue miR-21 levels were significantly associated with tumor size, distant metastasis, and reduced survival, marking it as an independent prognostic factor for CRC. An encompassing meta-analysis of 18 studies, which included 1,129 CRC patients, presented a sensitivity and specificity of circulating miR21 expression for CRC at 77% (95% confidence interval [CI], 70%–82%) and 83% (95% CI, 78%–88%), respectively. These findings hint at its considerable diagnostic value for CRC, marked by moderate sensitivity and good specificity [110]. Nevertheless, it is worth noting that miR-21 might also serve as a biomarker for other cancers (like breast, pancreas, lung, and stomach) or non-malignant conditions, indicating its broader role as a general disease marker [111,112]. miR-92a, part of the miR-17-92a cluster, is known to be upregulated in CRC. Its involvement in CRC tumorigenesis, metastasis, and treatment response has been extensively explored [113]. One study that examined a panel of 95 miRNAs highlighted that miR-17-3p and miR-92 levels in plasma were notably higher in CRC patients. Using a cutoff value of 240, the sensitivity and specificity were found to be 89% and 70%, respectively. Furthermore, miR-92a was able to distinguish CRC from other gastrointestinal cancers and inflammatory bowel disease [114]. A subsequent study from the same group indicated the diagnostic value of both miR-29a and miR-92a for CRC and advanced adenoma. Plasma miR-92a was able to differentiate advanced adenoma from controls with a sensitivity of 64.9% and a specificity of 81.4%. Meanwhile, miR-29a differentiated advanced adenoma from controls with a sensitivity of 62.2% and a specificity of 84.7%. Additionally, miR-29a expression was significantly higher in CRC compared to adenoma and was correlated with more advanced TNM stages [115]. In a meta-analysis encompassing 6 studies with 521 CRC patients, the sensitivity, specificity, and diagnostic odds ratio (DOR) for predicting CRC patients using miR-92a were 76% (95% CI, 72%–79%), 64% (95% CI, 59%–69%), and 8.05 (95% CI, 3.50–18.56), respectively. Moreover, the area under the curve (AUC) for miR-92a in diagnosing CRC was recorded at 0.7720 [116]. Subsequent meta-analyses have examined the role of individual miRNAs as CRC diagnostic biomarkers. One such analysis, which involved 16 studies, found miR-31 expression to be associated with diminished overall survival (OS) (hazard ratio [HR], 0.68; 95% CI, 0.47–0.97) and progression-free survival (HR, 0.49; 95% CI, 0.33–0.73). This miRNA also showcased significant predictive value for responses to anti-epidermal growth factor receptor (EGFR) treatment [117]. Another meta-analysis on miR-20a as a CRC biomarker revealed that its expression levels, whether in stool, serum, or tumor tissue, were notably higher in CRC patients versus controls. The pooled area under the receiver operating characteristic curve was determined to be 0.70, which is comparable to those of carcinoembryonic antigen and carbohydrate antigen 19-9, suggesting miR-20a may be useful CRC diagnostic biomarker [118]. Carter et al. [119] conducted a systematic review and meta-analysis of 34 studies that evaluated plasma or serum miRNA in diagnosing CRC. The aggregated results indicated that the overall sensitivity and specificity of 28 individual miRNAs stood at 76% (95% CI, 72%–80%) for both. This points to the substantial ability of miRNAs to act as noninvasive blood-based biomarkers for CRC detection. A further meta-analysis, which incorporated 35 studies with 3,258 CRC patients and 2,683 healthy participants, presented the result that single miRNAs had a sensitivity and specificity of 80% (95% CI, 75%–83%) and 80% (95% CI, 75%–84%) respectively, in CRC diagnosis. The positive likelihood ratio was 4.0 (95% CI, 3.2–5.0), the negative likelihood ratio was 0.26 (95% CI, 0.21–0.31), and the DOR was 16 (95% CI, 11–23). The AUC was computed as 0.87 (95% CI, 0.83%–0.89%). Furthermore, the findings revealed that miRNAs derived from serum samples distinguished CRC patients from controls with the highest precision, especially when placed with other biological samples [120].
The combination of miRNA into a biomarker panel has advanced with the progression of high-throughput microarray and sequencing technologies, and their clinical significance in pinpointing early CRC, and new therapeutic targets has been assessed. However, the diagnostic precision of integrating multiple miRNAs in CRC remains uneven, largely because many studies have sampled a relatively small patient count, often fewer than 100 [121]. A recent investigation that assessed serum miRNA expression from 85 CRC patients and 78 HCs indicated that serum levels of 5 miRNAs (miR-21, miR-29a, miR-92a, miR-125b, and miR-223) were considerably elevated in CRC patients. When combined, these miRNAs presented an AUC of 0.952, with a sensitivity of 84.7% and a specificity of 98.7% [122]. Another study, examining the predictive capability of serum miRNAs in a community-based sample (97 CRC cases and 103 frequency-matched HCs), found that 3 miRNAs (miRNA-29a, miRNA-125b, and miRNA-145) were substantially linked with incident CRC risk. The sensitivity of these 3 miRNAs ranged between 0.854 and 0.961. The basic model’s AUC, which only included basic demographic information, rose from 0.61 to 0.71 upon the addition of these 3 miRNAs [123]. Basati et al. [124] observed that serum levels of miR-194 and miR-29b, both of which are downregulated in CRC, were significantly diminished in CRC patients compared to HCs. These levels were inversely associated with advanced tumor stages and unfavorable outcomes, hinting at their potential as diagnostic and prognostic biomarkers for CRC. Peng et al. [125] noted that out of 96 irregularly expressed miRNAs identified via realtime polymerase chain reaction, miRNA-378* and miRNA-145 were notably downregulated in CRC tumor tissues, suggesting their potential as early CRC detection biomarkers. A meta-analysis spanning 20 studies with 3,339 CRC patients and 2,468 HCs revealed that miRNA panels for CRC diagnosis had 85% (95% CI, 84%–86%) sensitivity and 79% (95% CI, 78%–80%) specificity. Serum samples, when compared with other sample types, demonstrated the best diagnostic accuracy in subgroup analyses [126]. Notably, the global prevalence of early-onset CRC (EOCRC) in individuals younger than 50 is on the rise. EOCRC tends to have less favorable survival outcomes than its late-onset counterpart. A recent research study unearthed a novel liquid biopsy miRNA signature comprising 4 miRNAs (miR-193a-5p, miR-210, miR-513a-5p, and miR-628-3p) from blood samples of 72 EOCRC patients and 45 control subjects in Japan. In a validation set featuring 77 EOCRC patients and 45 control subjects from Spain, this 4-miRNA panel discerned EOCRC patients with an AUC of 0.92 for stage I/II CRC and 0.87 for stage III/IV CRC, implying its utility in detecting early-stage EOCRC [127]. Table 4 summarizes the most promising miRNAs (and miRNA panels) as potential blood-based noninvasive diagnostic biomarkers for CRC.

List of Most Promising miRNAs and miRNA Panels as Potential Blood-Based Noninvasive Diagnostic Biomarkers for CRC
Exosomes are extracellular vesicles secreted by most cell types, including cancer cells, into body fluids. They have a pivotal role in intracellular communication, cell signaling, tumor development and metastasis, as well as immune responses [128]. Exosomes produced by cancer cells are known to contain miRNA, and specific exosomal miRNAs offer potential as novel biomarkers for the early detection, prognosis, and treatment prediction of CRC [129]. Ogata-Kawata et al. [130] identified a panel of serum exosomal miRNAs (let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, and miR-23a) as promising biomarkers for CRC detection. Their expression levels in serum were considerably higher in patients with primary CRC, including those in early disease stages, compared to control individuals. Another research study noted that a set of 6 circulating exosomal miRNAs (miR-19a, miR-20a, miR-143, miR-145, miR150, and let-7a) in serum were significantly elevated in CRC patients, highlighting their potential as diagnostic biomarkers for CRC [131].
2. Prognostic and Predictive miRNA Biomarkers
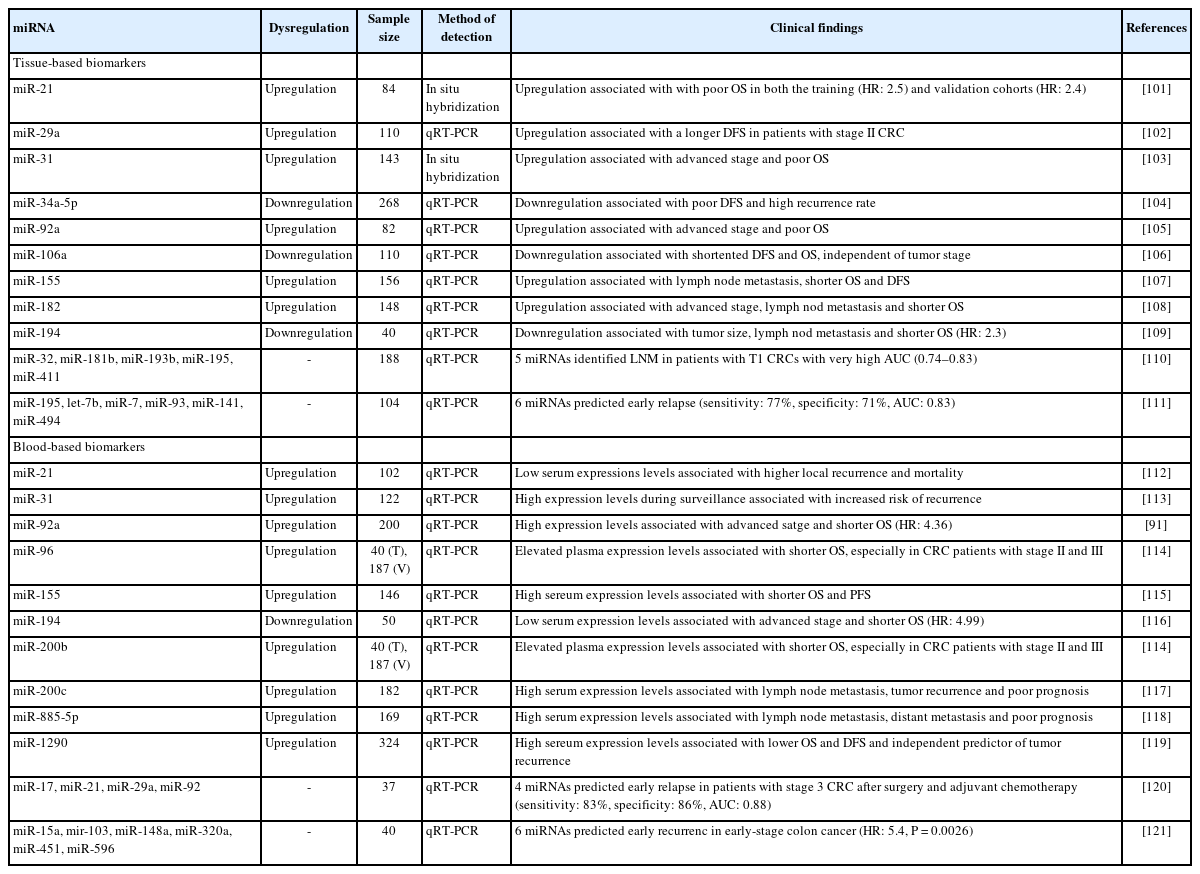
The potential of individual miRNAs or miRNA panels as prognostic or predictive biomarkers for CRC patients has been extensively researched. The pioneering study by Schetter et al. [108] found that 37 miRNAs were differentially expressed in CRC tissues. Among these, 5 miRNAs (miR-20a, miR-21, miR-106a, miR-181b, and miR-21) had notably higher expression levels in CRC tissues than in corresponding non-tumorous tissues, and this was validated in a Western cohort. Elevated miR-21 expression was linked to poorer survival, independently of clinical and pathological parameters, as well as adverse therapeutic outcomes in the test cohort. Furthermore, a validation study confirmed the significant association between heightened miR-21 expression and poorer survival in an Asian CRC cohort. Wang et al. [132] showed that miR-31 expression was markedly higher in CRC tissues compared to normal mucosa. This expression correlated positively with advanced TNM stages and deeper tumor invasion, suggesting a connection between miR-31 overexpression and CRC onset and progression. Table 5 summarizes most promising miRNAs (and miRNA panels) as potential prognostic biomarkers for CRC. Kjersem et al. [133] evaluated miRNA expression in plasma samples from 24 metastatic CRC patients, discovering that 3 miRNAs (miR-106a, miR-484, and miR-130b) were significantly more expressed in non-responders than in responders to oxaliplatin-based treatments. Gherman et al. [134] reported that elevated exosomal expression of miR-92a-3p and miR-221-3p might indicate resistance to first-line chemotherapy and was linked to shorter OS. A recent review highlighted various miRNAs that potentially influence 5-fluorouracil resistance, including miR10b, miR-19b, miR-20a, miR-21, miR-23a, miR-31, miR-34, miR-129, miR-140, miR-145, miR-192, miR-215, the miR-200 family, and miR-451 [1]. Nevertheless, most of these findings stem from preclinical research, underscoring the need for more extensive clinical studies for validation.
3. Stool-Based MicroRNA Biomarkers
Fecal miRNAs are stable, retaining a significant portion of their original level for up to 72 hours at room temperature. This stability underscores the potential for stool-based miRNA as a noninvasive detection method for CRC. Furthermore, repeated sampling of the same specimen has shown consistent results, indicating the high reproducibility of fecal miRNA detection [135,136]. In 2009, Ahmed et al. [137] became the first to report the detection of specific miRNAs in the stool samples of patients with sporadic colon cancer. Their findings revealed that 7 miRNAs (miR-21, miR-106a, miR-96, miR-203, miR-20a, miR-326, and miR-92) were upregulated, while 7 others (miR-320, miR126, miR-484-5p, miR-143, miR-145, miR-16, and miR-125b) were downregulated. Notably, the expression of the upregulated miRNAs was more prominent in later Dukes’ stages than in adenomas. The miRNA gene expression profile could distinguish between patients with CRC and those with active ulcerative colitis. In a subsequent study involving 197 CRC patients and 134 HCs, it was observed that fecal expression of the miR17-92 cluster and miR-135 was significantly elevated in CRC patients. The overall sensitivity and specificity stood at 74.1% and 79.0%, respectively. Notably, the sensitivity of cancer detection based on tumor location was significantly greater in distal CRC than in proximal CRC [138].
Several studies have reported miRNA panels with sensitivities exceeding 80% for CRC detection [135,139,140]. Wu et al. [135] examined the diagnostic accuracy of stool-based miRNA for both advanced adenoma and CRC. Their findings indicated that the expression of miR-21 and miR-92a was notably elevated in tissues and stools of CRC patients in comparison with controls. However, only fecal miR-92a levels were significantly higher in adenoma patients than in controls. The overall sensitivity of fecal miR-92a was 71.6% for CRC and 56.1% for adenoma, with a specificity of 73.3%. Moreover, fecal miR-92a displayed a greater sensitivity for distal CRC compared to proximal CRC, and a higher sensitivity for advanced adenomas over non-advanced ones. Following CRC or advanced adenoma treatment, fecal miR-92a levels were reduced. In research conducted by our group, CRC-related miRNAs were analyzed in stool samples from 29 CRC patients and 29 controls. Out of the 8 miRNAs tested, miR-21, miR-92a, miR-144*, and miR-17- 3p showed significantly elevated levels in the CRC cohort. The sensitivities and specificities of miR-21, miR-92, miR-144*, and miR-17-3p were 79.3% and 48.3%, 89.7%, and 51.7%, 78.6% and 66.7%, and 67.9% and 70.8%, respectively. Multivariate analysis indicated that miR-92a and miR-144* were strongly linked with the presence of CRC, highlighting their potential as noninvasive CRC biomarkers [139]. Zhu et al. [141] found that miR-29a, miR-223, and miR-224 levels in the stool of CRC patients were markedly lower than in healthy volunteers, suggesting that this miRNA panel might serve as a valuable tool for CRC screening and early detection. Duran-Sanchon et al. [140] formulated and verified a fecal miRNA-based algorithm that encompassed 2 upregulated CRC fecal miRNAs (miR-421 and miR-27a-3p), combined with hemoglobin concentrations, age, and gender of FIT-positive individuals. This combination identified CRC patients with an AUC of 0.93, contrasting with the AUC of 0.67 for FIT alone. However, its efficiency dropped to an AUC of 0.70 when patients with advanced adenoma were included. This algorithm was also capable of distinguishing CRC patients from those with non-advanced adenomas or those having a negative colonoscopy result, achieving an AUC of 0.9 and potentially avoiding 34% of colonoscopies [142]. Such findings suggest that the accuracy of the fecal miRNA-based algorithm could surpass that of FIT alone, potentially enhancing the effectiveness and efficiency of FIT-based CRC screening initiatives. A recent systematic review examined 20 studies focusing on 31 individual miRNAs and 16 miRNA panels for CRC detection. The reported diagnostic performance displayed a wide array of values, with AUCs ranging from 0.64 to 0.97, sensitivities between 15% and 97%, and specificities spanning 38% to 100%. Out of the 31 miRNAs, 10, including miR-21, miR-92a, miR-20a, miR-223, miR-144-5p, miR-135b, miR-18a, miR-29a, miR-451, and miR-221, were significantly linked with CRC in at least 2 studies. miR-21 was the most frequently mentioned miRNA across 5 studies, often appearing in miRNA panels [143]. A more recent study undertook comprehensive miRNA profiling using small RNA sequencing in stool samples, aimed at differentiating CRC patients from control subjects and identifying premalignant lesions. Among 25 miRNAs exhibiting altered profiles in the stool of CRC patients from 2 distinct European cohorts, 5 miRNAs (miR-149-3p, miR-607-5p, miR-1246, miR-4488, and miR-6777-5p) distinguished CRC patients from controls with an AUC of 0.86. Moreover, these miRNA profiles could accurately categorize patients with low-/high-stage tumors and advanced adenoma, compared to controls, with an AUC of 0.82 [144]. Koga et al. [145] assessed the potential of fecal miRNA for CRC detection using leftover stool samples from previous FIT procedures. The combined sensitivity and specificity of miR-106a with FIT were 70.9% and 96.3%, respectively, outperforming FIT alone (61% sensitivity and 98% specificity). Table 6 summarizes the most promising miRNAs (and miRNA panels) as potential stool-based noninvasive diagnostic biomarkers for CRC.
LONG NONCODING RNAs
Long noncoding RNAs (lncRNAs) are transcripts exceeding 200 nucleotides in length that lack protein-coding abilities, yet can undergo processing similar to miRNAs. Found abundantly in the human body, lncRNAs regulate gene expression by interacting with DNA, RNA, and proteins, serving various functional roles. The precise count of functional lncRNAs remains ambiguous as novel lncRNAs are continuously identified, and their roles have not been fully elucidated [121]. Over the past decade, mounting evidence suggests that lncRNAs exhibit oncogenic roles through epigenetic modifications, autophagy regulation, tumor microenvironment adjustments, and stem phenotype enhancement. Detected in blood, lncRNAs have emerged as potential biomarkers for both the diagnosis and prognosis of CRC, given their involvement in CRC’s pathogenesis and regulation [146].
1. Diagnostic Biomarkers
Numerous studies have spotlighted differentially expressed lncRNAs in CRC tissues in comparison with normal tissues. Furthermore, these expression levels have been associated with the clinicopathological features of the disease [147,148]. Among these, the most extensively researched oncogenic lncRNAs in CRC include HOX transcript antisense intergenic RNA (HOTAIR), metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), colorectal cancer-associated transcript 1 (CCAT1), and MIR31HG [148]. HOTAIR is a 2158-bp gene situated at the mammalian HOXC locus on chromosome 12q13.13, and is transcribed opposite to the HOXC gene. Expression levels of HOTAIR are elevated in CRC tissues compared to their normal counterparts and show close ties with the PRC2 complex (comprising SUZ12, EZH2, and H3K27me3), as identified by gene set enrichment analysis using complementary DNA array data [149]. Svoboda et al. [150] highlighted that HOTAIR expression levels were notably higher in both primary tumors and blood of CRC patients, encompassing those with early-stage CRC. This was associated with a less favorable prognosis, indicating its potential as both a diagnostic and prognostic biomarker. CCATs represent a set of lncRNAs observed to be upregulated in CRC and are found on chromosome 8q24, a region commonly amplified across various cancer types, including CRC [151]. Likewise, CCAT1 expression levels are significantly elevated in both primary tumors and blood, suggesting its role as an early marker of CRC development. Moreover, in addition to CCAT1, CCAT2 is identified at elevated levels across all stages of colon cancer. The high expression of these lncRNAs, whether individually or in tandem, in tumor tissue from CRC patients, was significantly linked to poorer recurrence-free survival and OS rates. This suggests their potential as biomarkers for both the diagnosis and prognosis of CRC [152]. Zhao et al. [153] assessed the diagnostic performance of 13 cancer-related lncRNAs. They found that plasma levels of CCAT1 and HOTAIR were notably higher in CRC patients than in HCs. Receiver operating characteristics curve analysis revealed the AUC for CRC detection was approximately 0.836 for CCAT1 and 0.777 for HOTAIR. When combined, these 2 lncRNAs displayed strong diagnostic capabilities for CRC screening, particularly in early-stage CRC, achieving an AUC of 0.954 with a sensitivity of 84.3% and specificity of 80.2%. Another study pinpointed 11 differentially expressed lncRNAs in CRC versus normal tissues. LINC01485 was found to be upregulated in CRC tissues compared to surrounding non-tumor tissues. In the CRC group, whole blood LINC01485 expression surged, showcasing a sensitivity and specificity of 98.33% and 84.00% respectively in distinguishing CRC from HCs [154]. Tao et al. [155] detailed that urothelial carcinoma associated 1 (UCA1) was substantially upregulated in colon cancer tissues in comparison to adjacent non-tumor tissues. Elevated UCA1 expression correlated with advanced tumor stages and a worse prognosis. Moreover, plasma UCA1 levels in colon cancer patients were significantly greater than those in HCs but decreased post-surgery, suggesting its potential as a biomarker for early diagnosis and disease tracking of colon cancer. Other lncRNAs like FLANC [156], MIR17HG [157], SNHG5 [158], and NEAT1 [159], have also been identified as potential diagnostic biomarkers.
A recent case-control study identified 3 novel lncRNAs, XLOC_006844, LOC152578, and XLOC_000303, using highthroughput lncRNA microarray. These were found to be upregulated in CRC patients when compared to HCs, with AUCs of 0.919 and 0.975 in the training and validation sets respectively, hinting at their potential as a biomarker panel for CRC detection [160]. Gharib et al. [161] examined the levels of a panel of 10 significantly dysregulated lncRNAs (CCAT1, CCAT2, H19, HOTAIR, HULC, MALAT1, PCAT1, MEG3, PTENP1, and TUSC7) identified in stool samples from 150 CRC patients. The diagnostic performance of this panel for differentiating CRCs from HCs showed an AUC of 0.8554 in the training set and 0.8465 in the validation set for all CRC stages (I‐IV TNM stages). Specifically, for early CRCs (I‐II TNM stages), the AUC values were consistent at 0.8554 for the training set and 0.8465 for the validation set. For advanced CRCs (III‐IV TNM stages), the AUC was 0.9281 in the training set and 0.9236 in the validation set. These data suggest the potential efficacy of stool lncRNAs for CRC screening. In summary, lncRNAs have risen to prominence as potential biomarkers for CRC detection. However, more comprehensive studies are essential to decipher the roles of oncogenic lncRNAs in CRC carcinogenesis and to affirm their status as biomarkers across a broader patient cohort.
2. Prognostic and Predictive Biomarkers
Dysregulation of lncRNAs expression in tumor tissue and blood can be associated with poor prognosis, and factors such as recurrence-free survival, OS, metastasis, tumor stage, or grade, highlighting their potential as prognostic and predictive biomarkers. MALAT1, one of the most abundantly expressed lncRNAs in human cells, has been identified as a prognostic biomarker in stage I non-small cell lung cancer [162]. Zheng et al. [163] found that MALAT1 expression was significantly elevated in stage II/III CRC tissues compared to non-tumor tissues, and its high expression was linked to adverse outcomes, including shorter disease-free survival and OS. A recent study examining several oncogenic lncRNAs in blood samples from 63 CRC patients and 40 HCs revealed elevated expression levels of MALAT1, CCAT1, and PANDAR compared to HCs, suggesting their potential as CRC prognostic biomarkers [164]. High expression levels of HOTAIR have been correlated with advanced tumor stage, lymph node metastasis, and unfavorable prognosis in CRC patients. A meta-analysis of 6 studies found that elevated HOTAIR expression predicted poorer OS and recurrencefree survival in CRC patients and was significantly linked to venous invasion, advanced tumor infiltration, and distant metastasis [165]. Another study analyzed a broad panel of lncRNAs in a CRC dataset from The Cancer Genome Atlas (TCGA) and identified H19 as the most significant lncRNA linked to shorter OS in CRC patients, a finding validated in 2 separate CRC cohorts [166]. HOXA transcript at the distal tip (HOTTIP) lncRNA expression was notably higher in CRC tissues than in non-tumor tissues and was associated with tumor stage and distant metastasis in CRC patients [167]. RP11 was found to be highly expressed in CRC tissues, correlating with advanced CRC stage and poor prognosis. The biological functions of RP11 include promoting the migration, invasion, and epithelial-mesenchymal transition of CRC cells in vitro and enhancing liver metastasis in vivo [168]. In summary, lncRNAs hold promise as noninvasive biomarkers for CRC prognosis, treatment, and diagnosis. Still, further investigations are essential to confirm their clinical relevance and define their role in disease management.
CONCLUSIONS
Epigenetic modifications are common in colorectal premalignant lesions and cancers. There is growing evidence suggesting the potential of aberrant DNA methylation and alterations in ncRNAs as diagnostic and prognostic biomarkers for CRC. Notably, the FDA has approved a stool DNA test using a multi-target panel that includes DNA methylation, and updated guidelines now recommend this test as a CRC screening method. Ongoing research and advancements in molecular techniques to identify epigenetic alterations as biomarkers for early detection, prognostication, and treatment prediction are vital for managing CRC patients. Moreover, larger randomized controlled studies are needed to validate epigenetic alterations in various biological fluids as tools for CRC screening, alongside technical enhancements for detecting specific epigenetic changes.
Notes
Funding Source
The authors received no financial support for the research, authorship, and/or publication of this article.
Conflict of Interest
No potential conflict of interest relevant to this article was reported.
Data Availability Statement
Not applicable.
Author Contributions
Writing and approval of the final manuscript: Oh CK and Cho YS.