Gut microbiota in pathophysiology, diagnosis, and therapeutics of inflammatory bowel disease
Article information
Abstract
Inflammatory bowel disease (IBD) is a multifactorial disease, which is thought to be an interplay between genetic, environment, microbiota, and immune-mediated factors. Dysbiosis in the gut microbial composition, caused by antibiotics and diet, is closely related to the initiation and progression of IBD. Differences in gut microbiota composition between IBD patients and healthy individuals have been found, with reduced biodiversity of commensal microbes and colonization of opportunistic microbes in IBD patients. Gut microbiota can, therefore, potentially be used for diagnosing and prognosticating IBD, and predicting its treatment response. Currently, there are no curative therapies for IBD. Microbiota-based interventions, including probiotics, prebiotics, synbiotics, and fecal microbiota transplantation, have been recognized as promising therapeutic strategies. Clinical studies and studies done in animal models have provided sufficient evidence that microbiota-based interventions may improve inflammation, the remission rate, and microscopic aspects of IBD. Further studies are required to better understand the mechanisms of action of such interventions. This will help in enhancing their effectiveness and developing personalized therapies. The present review summarizes the relationship between gut microbiota and IBD immunopathogenesis. It also discusses the use of gut microbiota as a noninvasive biomarker and potential therapeutic option.
INTRODUCTION
Inflammatory bowel disease (IBD) is characterized by chronic and relapsing inflammation of the gastrointestinal tract [1]. It encompasses 2 main clinical entities, Crohn’s disease (CD) and ulcerative colitis (UC). Though CD and UC show similar clinical symptoms, they vary in their anatomical distribution, which can occur anywhere in the gastrointestinal tract for CD but only in the colon and rectum for UC [2]. Depending on symptoms and disease severity, UC and CD are classified into mild, moderate, and severe [3]. IBD affects about 6.8 million people globally [4]. The incidence of IBD has been rising in developing countries [4,5], whereas a stabilizing trend has been observed in high-prevalence developed countries in Europe and North America [6]. Individuals with IBD are at greater risk of developing colorectal cancer (CRC), called colitis-associated cancer, than normal individuals [7-9]. The risk of developing CRC is about 20% and 2.5% to 4.5% for patients with UC [10] and CD [11], respectively. Colitis stimulates carcinogenesis by inducing the expansion of genotoxic bacteria [12], Patients with IBD show a significant clinical heterogeneity, which makes the right treatment for each patient difficult [13,14].
The human gastrointestinal tract contains a diverse assembly of microorganisms, including bacteria, archaea, fungi, protozoa, and viruses [15]. It has been estimated that there are approximately 40 trillion microorganisms that constitute the human gut microbiota [16]. The gut microbiota possesses approximately 150-fold more genes than the human genes [17]. The gut microbiota and humans show a symbiotic relationship, and the composition of the human gut microbiota is closely linked to health and disease [18,19]. Many factors influence the composition of gut microbiota, including the delivery (C-section or normal), diet, age, race, environmental factors, use of antibiotics, or population geography [20-28]. The gut microbiota constantly crosstalk with each other and the host cells and maintain the homeostasis of the intestinal environment.
More than 99% of the bacteria in the human intestine belong to the 4 major phyla: Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria [29,30]. Nearly 90% of bacterial phyla in the colon are represented by Bacteroidetes and Actinobacteria, while 40% of the bacterial phyla in the small intestine are represented by Firmicutes [26,31]. The composition and diversity of gut microbiota vary along the gastrointestinal tract and between mucosal surfaces and the lumen [32-35]. It has been found that the Firmicutes/Bacteroidetes ratio is a critical parameter for gut health [36]. Various studies have found that a change in the Firmicutes/Bacteroidetes ratio is associated with IBD [37-41].
Pathogenic and symbiotic microbiota can coexist in a healthy human gut. Any disturbance can alter the normal interactions between microbiota and the host, potentially affecting the susceptibility to IBD [33]. Such disturbance or change in microbiota, commonly referred to as “dysbiosis,” is involved in the pathophysiology of gastrointestinal diseases such as IBD and irritable bowel syndrome (IBS) [42-44]. A significant reduction in the complexity of the gut microbial ecosystem and alterations in a few specific taxa have been observed in patients with IBD [45]. The association of gut microbiota with IBD is strengthened by the fact that when germ-free mice are inoculated with IBD microbiota, they develop severe colitis [46,47]. However, it remains unclear whether dysbiosis of the gut microbiota is a cause or a consequence of IBD. An increase in microbial species capable of tolerating oxidative stress indicates that dysbiosis in IBD is caused by inflammation [48]. Though the precise etiopathology of the disease remains to be elucidated, one of the hypotheses suggests that an inappropriate immune response to the intestinal microbiota in genetically predisposed individuals can lead to IBD [49]. Over 200 IBD-associated susceptible genes have been identified, some of which are involved in host innate or adaptive immune responses to gut microbiota [3,50-52]. This indicates that gut microbiota play an essential role in the pathogenesis of IBD [53].
This review discusses the relationship between the gut microbiota and the onset and progression of IBD and their potential role in the diagnosis, treatment, and prevention of IBD.
GUT MICROBIOTA AND IBD
Various molecular techniques have been employed to study the human gut microbiota. These techniques have their own biases and limitations, and no known molecular technique is able to capture all of the microorganisms that inhabit the human gut. Moreover, as mentioned earlier, individual gut microbiota is affected by numerous factors, and comparing microbiota from different individuals is not often conclusive. However, it is still possible to reach a consensus by comparing different studies conducted using different techniques and experimental settings. By comparing cases and controls, we can identify recurring patterns of trends that provide a comprehensive understanding of the gut microbiota. Collectively they can contribute to a more robuse and reliable consensus regarding the characteristics of the gut microbiota in diseases.
Studies have shown that alterations in the gut microbiota, often called dysbiosis, are associated with IBD [54,55]. Burgeoning evidence suggests that a reduction in fecal microbial diversity is an indicator of IBD. A decline in Firmicutes and Bacteriodetes [42,43,53,56-58] and an increase in Proteobacteria and Actinobacteria have been observed in IBD [54,58,59]. Specific genera, such as Bacteroides, Eubacterium, Faecalibacterium, and Ruminococcus, are reduced in fecal samples of patients with CD [60]. Nemoto et al. [61] observed that patients with UC showed a decline in Bacteroides and Clostridium XIVab. In another study, Fuentes et al. [62] found that patients with UC had fewer Clostridium and increased Bacteriodetes, whereas Khalil et al. [63] found that sulfate-reducing bacteria were dominant in UC but were fewer in healthy subjects. A multicenter study by Gevers et al. [64] reported an increase in the members of Pasteurellaceae, Veillonellaceae, and Enterobacteriaceae and a decrease in the members of Clostridioides, Bacteroidales, and Erysipelotrichales in CD patients. Members of the family Enterobacteriaceae express lipopolysaccharide that can trigger strong inflammatory responses, contributing to IBD [65,66]. Studies have also found decreased abundance of Clostridium lavalense, Ruminococcus torques, Blautia faecis, and Roseburia inulinivorans in patients with CD [67,68]. Takahashi et al. [68] reported a decrease in Faecalibacterium, Eubacterium, Bacteroides, and Ruminococcus and an increase in Actinomyces and Bifidobacterium in CD patients.
In addition to changes in bacterial diversity, changes in archaeal [55,69-71], and viral diversity [72] have also been reported in IBD. Interkingdom crosstalk between the bacteriome and mycobiome has been implicated in CD.73
1. IBD-Associated Bacteria
A change in the human bacteriome has been found in IBD (Table 1), due to their greater abundance than other kingdoms of microbes in the gut. The bacteria found to be associated with IBD include Escherichia coli, Bacteroides fragilis, Ruminococcus gnavus, Faecalibacterium prausnitzii, and Roseburia (Fig. 1).

Microbes involved in inflammatory bowel disease and their molecular mechanisms. Figure created with BioRender. B. fragilis, Bacteroides fragilis; E. faecium, Enterococcus faecium; E. coli, Escherichia coli; F. nucleatum, Fusobacterium nucleatum; F. prausnitzii, Faecalibacterium prausnitzii; M. stadtmanae, Methanosphaera stadtmanae; R. gnavus, Ruminococcus gnavus; TLR4, Toll-like receptor 4; BFT, B. fragilis toxin; TNF, tumor necrosis factor; ETBF, enterotoxigenic B. fragilis; AhR, aryl hydrocarbon receptor; IL, interleukin; PLC, phospholipase C; DAG, diacylglycerol; PKC, protein kinase C.
1) Escherichia coli
E. coli is a Gram-negative, facultative anaerobic bacterium and a normal inhabitant of the human gut. The gut microbiota of patients with IBD show an increased abundance of adherentinvasive E. coli (AIEC) [74-77]. AIEC can adhere to and cross the intestinal mucosa of IBD patients, induce inflammation, and increase the permeability of the intestinal epithelium [78,79]. Mancini et al. [80] found that AIEC could disrupt epithelial mitochondrial networks and influence gut permeability by affecting gene expression. AIEC can survive and replicate in macrophages, resulting in the secretion of tumor necrosis factor (TNF) and, thus, inflammation [81,82]. A high abundance of E. coli has been linked to inflammation in patients with IBD [83].
2) Bacteroides fragilis
B. fragilis is a commensal Gram-negative anaerobic bacterium. It is an opportunistic pathogen with pro-inflammatory properties [84] and contributes to IBD [85]. Strains of B. fragilis that express a zinc-dependent metalloprotease, called B. fragilis toxin (BFT or fragilysin), are known as enterotoxigenic B. fragilis (ETBF) [86,87]. The strains that lack BFT are called nontoxigenic B. fragilis (NTBF). ETBF is responsible for diarrhea in children [88] and IBD [89,90]. BFT directly affects signaling pathways such as Wnt, NF-κB, STAT3, and MAPK pathways, leading to the production of pro-inflammatory mediators [91-95]. ETBF activates the Stat3 transcription factor, increases Th17 and T regulatory cells (Treg), and promotes mucosal permeability [96,97]. ETBF also induces reactive oxygen species (ROS) production and DNA damage by inducing the expression of spermine oxidase in colonocytes [98]. Kordahi et al. [99] found a correlation between B. fragilis and the levels of inflammatory cytokines. Contrary to ETBF, NTBF can induce the anti-inflammatory effects of Treg and direct the development of CD4+ T cells [100,101].
3) Ruminococcus gnavus
R. gnavus is a strict anaerobic Gram-positive bacterium and a part of the normal intestinal flora in humans. An increased abundance of R. gnavus is associated with IBD [57,102-104]. Hall et al. [103] found that R. gnavus could reach an abundance of 69% in patients with severe CD. R. gnavus produces complex glucorhamnan polysaccharide, which can induce the secretion of inflammatory cytokines, like TNF-α, by dendritic cells [104].
4) Faecalibacterium prausnitzii
F. prausnitzii is one of the most important butyrate-producing bacteria found in the gastrointestinal tract [105,106], which has drawn much attention in recent years. A decrease in F. prausnitzii has been reported in IBD [67,107-114]. Varela et al. [115] found that UC patients had insufficient colonization of F. prausnitzii, and the maintenance of clinical remission required F. prausnitzii colonization. They also found that the abundance of F. prausnitzii was correlated with a relapse of ileal CD after surgery. F. prausnitzii mediates anti-inflammatory effects by inhibiting the NF-κB pathway in intestinal epithelial cells [111] and by producing butyrate, which maintains a balance of Th17/Treg cells [107,116]. F. prausnitzii also stimulates the production of anti-inflammatory cytokines, such as interleukin (IL)-10, and inhibits the production of inflammatory cytokines, such as IL-12 and interferon-γ [107].
5) Roseburia
Roseburia is a genus of anaerobic, Gram-positive, rod-shaped bacteria known for butyrate production in the human colon. Several studies have confirmed that a decrease in Roseburia is associated with IBD [109,113,117-119]. Roseburia intestinalis can convert acetate to butyrate and has anti-inflammatory effects [118]. The patients with high genetic risk for IBD have significantly reduced Roseburia spp [117]. Other bacteria, such as Enterococcus faecium, Citrobacter rodentium, and Mycobacterium, can promote cytokine expression and inflammation in the colon, inducing IBD [120-122]. Fusobacterium is also found to be abundant in the colonic mucosa of UC patients [123]. Fusobacterium nucleatum is a pro-inflammatory bacterium that activates epithelial TLR4, resulting in inflammation [124,125].
2. IBD-Associated Archaea
Archaea are single-celled prokaryotes like bacteria but are closer to eukaryotes at the genetic level. Like bacteria, they also inhabit various locations of the human body [126-129]. The most dominant archaea in the human gut are methanogens, particularly Methaonbrevibacter and Methanosphaera [126]. Studies have found that a decrease in methanogens like Methanobrevibacter smit-hii is associated with IBD [69-70]. M. smithii plays a role in digestive health by directing Bacteroides thetaiotaomicron-mediated fermentation of dietary fructans to acetate [130]. Contrary to M. smithii, studies have found that the abundance of Methanosphaera stadtmanae increases up to 3-fold in IBD patients [131,132]. M. stadtmanae stimulates pro-inflammatory cytokine production by dendritic cells [132]. In addition to methanogens, halophilic archaea may also be involved in the etiology of IBD [133]. Contrary to these results, Chehoud et al. [134] reported that there were no alterations in the archaeome associated with IBD. Nevertheless, their role in the onset and progression of IBD is still debatable [127], probably due to a lack of sufficient studies.
3. IBD-Associated Fungi
Fungi represent just 0.1% of the total microbial community in the gut [135]. Alterations in the mycobiome are also associated with IBD. Studies have found reduced fungal diversity and dysbiosis in patients with IBD [55,71,73,136]. Sokol et al. [55] found an increased Basidiomycota/Ascomycota ratio in patients with IBD. Other studies have reported an increase in Candida, Malasseziales, and Filobasidiaceae and a decrease in Saccharomyces, Penicillium, and Kluyveromyces [137,138]. Candida is a pathogenic fungus and has gathered much attention in recent years. Several studies have reported an increased abundance of Candida in IBD patients [73,134,139,140]. Studies have also reported the involvement of Candida in causing severe colitis in a mouse model [141,142]. Mounting evidence suggests that colonization by Candida albicans enhances inflammation by increasing IL-17 and IL-23 production [143-145].
Contrary to Candida, decreased abundance of Saccharomyces cerevisae is reported in patients with IBD [55,139]. Limon et al. [151]. S. cerevisae may prevent IBD by preventing AIEC from adhering to the inflamed intestinal mucosa [146] and inhibiting aspartyl proteases-2 and 6 from C. albicans, preventing its transformation into an invasive hyphal form [147]. Saccharomyces boulardii may also have a preventive role in IBD owing to its anti-inflammatory effects and protection against intestinal pathogens [148,149]. Increased abundances of Malassezia have been reported in patients with IBD [55,150,151]. Limon et al. [151] found that Malassezia restricta was enriched in the mucosa of patients with CD. Many Malassezia strains can synthesize indole compounds that act on the aryl hydrocarbon receptor (AhR) [152-154] and also regulate the production of inflammatory mediators [155].
4. IBD-Associated Viruses
The human gut virome includes mostly bacteriophages and eukaryotic viruses [156,157]. Alterations in the gut virome have also been reported in IBD [72,158-160]. IBD is also associated with the loss of the core phagome and increased abundance of temperate phages [160]. However, contrary to bacterial diversity, increased virome diversity has been reported in IBD patients [72,159,161,162], indicating the role of gut virome in bacterial dysbiosis [162]. In particular, the abundance of bacteriophages of the family Caudovirales was found to increase in patients with UC and CD [72,159,161,162]. Caudovirales may contribute to IBD by decreasing bacterial richness.
Studies have also found significantly increased phage populations in the gut biopsies of patients with IBD compared to controls [163,164]. Duerkop et al. [165] reported that bacteriophages Caudovirales and Podoviridae were enriched in a mouse model of colitis. Similarly, Seth et al. [166] found that increased viral diversity was correlated with gut dysbiosis and levels of pro-inflammatory cytokines in mouse models. Gogokhia et al. [167] found that an increase in bacteriophage abundance aggravated colitis in germ-free mice. Pérez-Brocal et al. [168] reported that bacteriophages that infect Alteromonadales and Clostridiales were enriched in IBD. In another study, Cornuault et al. [169] found a high proportion of F. prausnitzii temperate phages in IBD patients, which results in the depletion of F. prausnitzii. Contrary to the above studies, Galtier et al. [170] found that bacteriophages could significantly reduce the abundance of AIEC in a mouse model of dextran sulfate sodium (DSS)-induced colitis. Similarly, Yang et al. [171] found that enteric viruses could ameliorate gut inflammation by stimulating interferon-β production. In addition to bacteriophages, eukaryotic viruses may also trigger intestinal inflammation and contribute to IBD pathogenesis [158,159,164,172]. Studies have found an increased abundance of the eukaryotic viruses Pneumoviridae [159] and Hepadnaviridae [173] in UC patients and Herpesviridae in IBD patients [164]. Cadwell et al. [174] found that in mice with a risk gene for CD, Atg16L1HM, norovirus could induce intestinal pathologies. In a similar study, Bolsega et al. [175] found that murine norovirus could induce colitis in an Il10-deficient mouse model of IBD in a microbiota-dependent manner.
GUT MICROBIOTA AS POTENTIAL BIOMARKER FOR IBD DIAGNOSIS
The current methods of assessing IBD activity include measuring inflammation using biomarkers from plasma and feces, such as C-reactive protein and fecal calprotectin, which are not IBD-specific. Other methods, such as colonoscopy, can precisely predict IBD but have limited clinical application due to their invasiveness and risk of intestinal perforation. The use of gut microbiota as a diagnostic tool for IBD can reduce the frequency of such invasive procedures. Stool samples can be used to generate data through 16S rRNA sequencing, metagenomic sequencing, or metabolomic profiling. The presence or absence of specific gut microbiota can also help predict treatment response in IBD [176]. The use of microbial biomarkers for IBD diagnosis is a promising method that needs to be utilized in clinical practice.
Lopez-Siles et al. [177] confirmed that F. prausnitzii was a specific indicator of IBD. They also reported that the patients with IBD had a lower abundance of F. prausnitzii than IBS patients and healthy controls. Combining F. prausnitzii with E. coli could even distinguish colonic CD from extensive colitis. Prosberg et al. [178] also found that the patients with active CD and UC had a lower abundance of F. prausnitzii than those in remission. These studies confirm that F. prausnitzii may be a reliable biomarker for IBD. A significant decrease in Bifidobacterium has also been reported in IBD patients [179-182]. Contrary to this, other studies have found an increased abundance of Bifidobacterium in IBD patients compared to controls [68,183,184]. Further research is required in this direction to know the precise role of Bifidobacterium in IBD pathogenesis.
Using gut microbiota, it is even possible to distinguish healthy individuals from UC or CD patients with an accuracy of 93.2% and 89.5%, respectively [59]. Fukuda and Fujita [185] used TRFLP of feces samples for operational taxonomic unit (OTU) discriminant analysis. They could distinguish between patients with active UC and other groups, which include individuals with mild inflammation in the large intestine, no inflammation, consanguineous-healthy individuals, and non-consanguineous healthy individuals. He et al. [186] reported a higher abundance of Klebsiella, Enterococcus, and Haemophilus in patients with active UC and a higher abundance of Roseburia, Lachnospira, Blautia, and Faecalibacterium in patients in remission. They also reported that decreased abundance of F. prausnitzii was related to a higher risk of recurrence in ileal CD. A combination of multiple microorganisms, including bacteria, archaea, and fungi, may serve as a more accurate and reliable biomarker to distinguish between individuals with UC, CD, and IBS from healthy individuals.
Gut microbiota can also be used as a biomarker for predicting the clinical relapse of IBD. A high abundance of Streptococcus has been reported in patients with postoperative recurrence [187]. Screening preoperative stool samples for Streptococcus may be a predictive marker of disease recurrence.
METABOLOMICS OF GUT MICROBIOTA IN IBD
The gut microbial metabolites include metabolites produced de novo by the gut microbes, metabolites produced by the host and modified by the gut microbes, and by-products of microbial interactions with dietary components. Metabolomics has been used to distinguish between IBD patients and healthy subjects [119,188-190]. Metabolites, particularly short-chain fatty acids (SCFAs), bile acids, and tryptophan metabolites, are found to be involved in IBD pathogenesis (Fig. 2). Various studies have confirmed reductions in the levels of medium-chain fatty acids and SCFAs [119,189,191,192], dysregulation of bile acid metabolism [113,193], and changes in the levels of amino acids [193,194] in the feces samples of IBD patients. Other studies have reported alterations in tricarboxylic acid cycle (TCA) intermediates [195,196], hippurate [197], and amino acid metabolism [198] in patients with IBD.
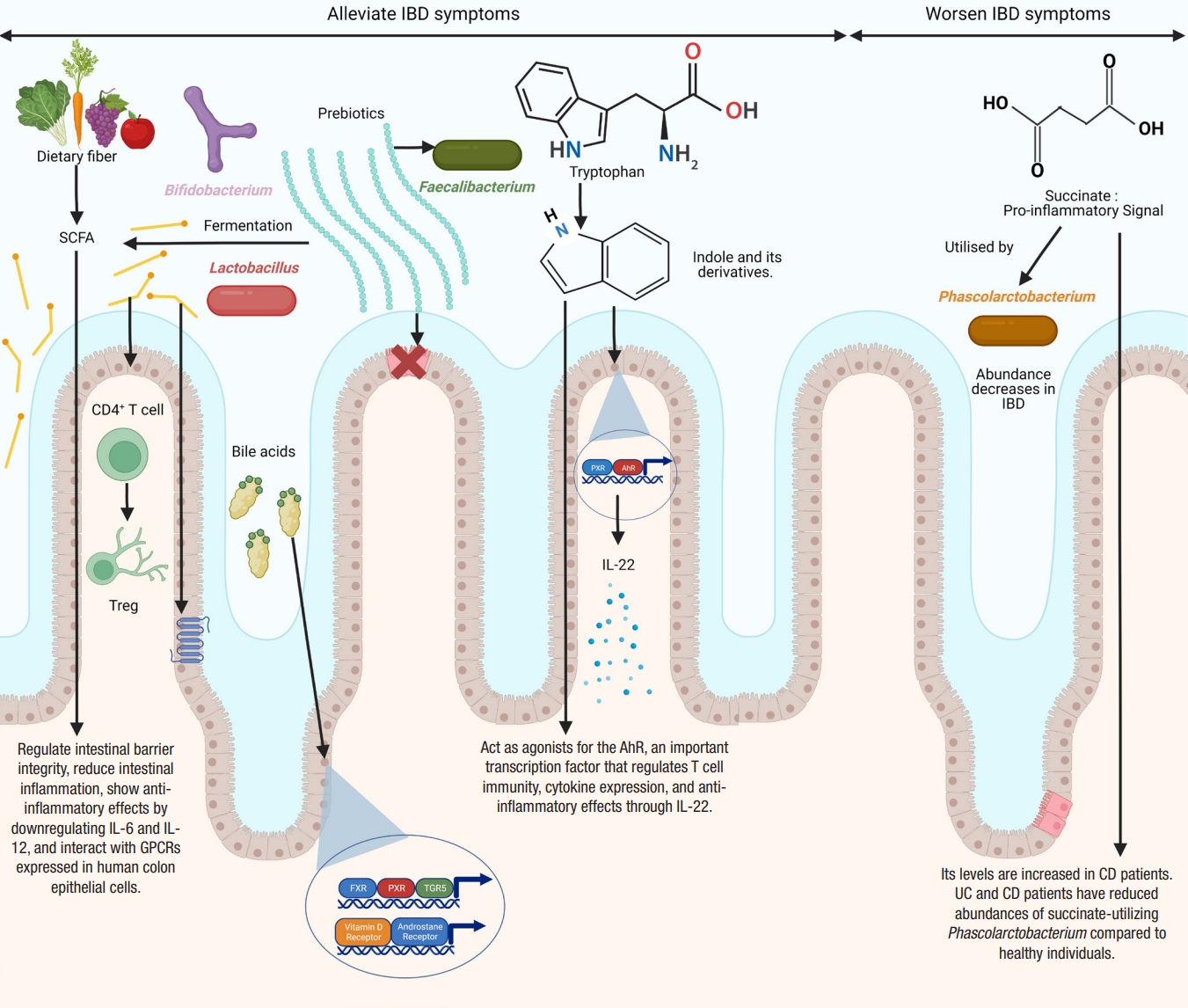
Microbial metabolites implicated in inflammatory bowel disease (IBD) and their molecular mechanisms. Figure created with BioRender. SCFA, short-chain fatty acid; PXR, pregnane X receptor; AhR, aryl hydrocarbon receptor; IL, interleukin; Treg, T regulatory; GPCRs, G protein-coupled receptors; CD, Crohn’s disease; UC, ulcerative colitis; FXR, farnesoid X receptor; TGR5, transmembrane G protein-coupled receptor 5.
Differences in the diversity and abundance of intestinal metabolites are seen in patients with IBD compared to control, with increased levels of primary bile acids, sphingolipids, and amino acids and decreased levels of indoles, long-chain fatty acids, cholesterol, and tetrapyrroles in IBD patients [119].
Studies have indicated that anti-inflammatory microbial metabolites are decreased [119,199], while pro-inflammatory microbial metabolites are enriched [118] in IBD patients compared to healthy subjects. Morgan et al. [200] reported that 12% of metabolic pathways significantly differed between IBD patients and healthy controls. They also found that the genes for the metabolism of butanoate and propanoate were decreased in CD patients.
1. Short-Chain Fatty Acids
SCFAs, such as acetate, propionate, and butyrate, are produced by the anaerobic fermentation of non-digestible carbohydrates by gut microbes [201-203]. Acetate accounts for nearly 50% to 70%, propionate for 10% to 20%, and butyrate for 20% to 30%. Acetate is produced by a variety of gut microbes [204], propionate is mainly produced by Bacteroidetes, Negativicutes, and Lachnospiraceae [201,205], while butyrate is mostly produced by Eubacterium, Clostridium, and Fusobacterium [206]. SCFAs are important players in regulating intestinal barrier integrity and reducing intestinal inflammation [109,207]. SCFAs regulate colonic Treg and show anti-inflammatory effects [208]. Butyrate and propionate can downregulate pro-inflammatory cytokines, such as IL-6 and IL-12 [209,210]. Butyrate and propionate can also alter the chromatin state by inhibiting histone deacetylases [211]. Furusawa et al. [212] found that butyrate upregulated histone H3 acetylation of the Foxp3 gene and promoted CD4+ T cell differentiation into Treg. Many studies have confirmed lower levels of SCFAs in the feces of IBD patients compared to healthy controls [109,113,213-215]. However, other studies have found a non-significant reduction in SCFAs in IBD [188,189]. Contrary to the levels of other SCFAs, lactate levels are elevated in both UC and CD [119,216,217].
SCFAs affect cellular functions by interacting with G protein-coupled receptors expressed in human colon epithelial cells [218]. Orphan G protein-coupled receptor (GPR43) interacts with acetate, propionate, and butyrate [219]. The binding of SCFAs with GPR43 reduces inflammation [100]. Loss of GPR43 results in refractory colitis in the DSS mice model [100]. Butyrate binds and activates GPR109A [220,221]. The interaction of butyrate with GPR109A reduces inflammation by promoting the differentiation of Treg, suppressing the induction of inflammatory cytokine, IL-6, and secreting anti-inflammatory cytokine, IL-10, by TH1 cells, and also activates macrophages and CD+ T cells [208,212,222,223]. SCFAs regulate intestinal homeostasis by stimulating the production of antimicrobial peptides and intestinal IgA [224,225]. They promote epithelial homeostasis through the production of IL-18 [226]. They also inhibit NF-κB expression and secretion of TNF-α [227]. They also have anti-proliferative effects [228] and a protective role in animal models of colitis [208] and colitis-induced CRC [221].
Butyrate is the major energy source for colonic epithelial cells and inhibits epithelial stem cells [229]. It modulates mitochondrial function, increasing oxygen consumption of colonic epithelial cells [230-233], which decreases oxygen concentration in the intestinal tract, increasing the number of obligate anaerobic bacteria, including butyrate-producing Firmicutes [234]. IBD is characterized by the loss of butyrate-producing bacterial species, such as F. prausnitzii, Roseburia hominis, and Clostridium cluster IV and XIVa, as is evident by a reduction in fecal butyrate levels in IBD patients [68,107,109,183]. Administration of butyrate by enema in UC has been found to be beneficial [235]. Therefore, decreased concentrations of SCFA-producing bacteria and SCFAs may be involved in chronic intestinal inflammation and the pathophysiology of IBD.
Studies have also found reduced levels of medium-chain fatty acids in IBD patients [189,236]. Medium-chain fatty acids may have an anti-inflammatory role, as replacing n-6 fatty acids with medium-chain fatty acids can reduce the incidence of colitis in IL-10-/- mice [237].
2. Bile Acids
Bile acids play an essential role in the emulsification and absorption of fats and the elimination of cholesterol. They are a type of steroid acid found in bile and synthesized from cholesterol by the liver. Primary bile acids, such as cholic acid and chenodeoxycholic acid, are synthesized in the host liver and are metabolized to secondary bile acids, such as deoxycholic acid and lithocholic acid by anaerobic microorganisms, particularly clostridial species, in the colon [238]. Many gut microbes can produce unique bile acids by conjugating amino acids to cholic acid [239]. Bile acids exert many metabolic effects by binding to various receptors, including the farnesoid X receptor (FXR), pregnane X receptor (PXR), transmembrane G protein-coupled receptor 5 (TGR5), vitamin D receptor, and androstane receptor [240].
A bidirectional relationship exists between bile acids and the microbiota. Gut microbiota can deconjugate amino acid residues from primary bile acids using the enzyme bile salt hydrolases [241]. Labbé et al. [242] analyzed metagenomics samples from the Human Microbiome Project and MetaHit. They found a reduction in the clusters of bile salt hydrolase (bsh) genes associated with Firmicutes in IBD. Bile acids have strong antimicrobial properties [243] and may change gut microbiota composition and density. They may have direct antimicrobial effects on bacteria such as Bifidobacterium breve and Lactobacillus salivarius [243]. The indirect antimicrobial effects of bile acids include stimulating the production of antimicrobial peptides from the host and activation of FXR [244-246]. Bile acids are crucial in the development of colonic RORγ+ Treg in mice, suggesting their anti-inflammatory effects [247].
Various studies have reported altered bile acid profiles in fecal samples of IBD patients [119,193,248-250]. Bile acid synthesis is regulated by the FXR. Once activated, FXR can reduce bile acid synthesis and uptake by downregulating gene expression in enterocytes [251]. FXR activation also exerts anti-inflammatory effects and is protective in chemically induced colitis [252,253]. Concomitant with this finding, studies have shown a significant reduction in FXR expression in CD patients [252,254].
3. Tryptophan Metabolites
Tryptophan is an essential aromatic amino acid. Alterations in the concentration of tryptophan and its metabolites and changes in the activity of associated enzymes have been reported in IBD [255]. Dietary tryptophan is metabolized by 3 metabolic pathways: the kynurenine pathway, serotonin pathway, and indole pathway. The indole pathway is the microbial pathway used by the gut microbes to metabolize indole, while the kynurenine and serotonin pathways are the endogenous pathways. The majority of dietary tryptophan is metabolized by the kynurenine pathway [256]. The microbial pathway can convert tryptophan into various bioactive indole derivatives, such as indole propionic acid, indole acetic acid, indole acrylic acid, indole 3-aldehyde, and tryptamine. These indole metabolites can act as agonists for the AhR, an important transcription factor that regulates T cell immunity, cytokine expression, and anti-inflammatory effects through IL-22 [257-261]. Concomitant with these findings, Monteleone et al. [262] reported that the inflamed mucosa of CD patients had reduced expression of AhR. Lactobacillus strains that can activate AhR have been shown to reduce the severity of DSS-induced colitis [263]. Indole metabolites can also activate PXR, which promotes intestinal barrier function in a mouse model of CRC [264]. Indole propionic acid inhibits TNF production by binding to PXR [258]. Reduced levels of indole propionic acid have been found in the serum of UC patients [265]. Other indole metabolites, such as indole acrylic acid, are also reduced in patients with IBD [258,266]. Wlodarska et al. [266] found that Peptostreptococcus russellii, a mucin-utilizing bacterium, could metabolize tryptophan to indole acrylic acid thus could reduce susceptibility to colitis. A critical study by Nikolaus et al. [255] identified a crucial link between tryptophan metabolism and IBD in a large clinical cohort of 535 patients. This study found an inverse relationship between tryptophan levels and IBD. A similar study found that a deficiency of dietary tryptophan could worsen colitis in mouse models [267].
4. Succinate
Succinate is a TCA intermediate produced by both the host and the microbiota [268]. Succinate metabolism has gathered much attention in recent years due to its potential link with IBD. Succinate acts as a key pro-inflammatory signal, and its levels are increased in CD [269,270]. Similarly, the levels of fecal succinate have also been found to be increased in UC and CD patients [216]. Concomitant with these findings, both UC and CD patients have reduced abundances of succinate-utilizing Phascolarctobacterium compared to healthy individuals [200].
GUT MICROBIOTA IN IBD THERAPEUTICS
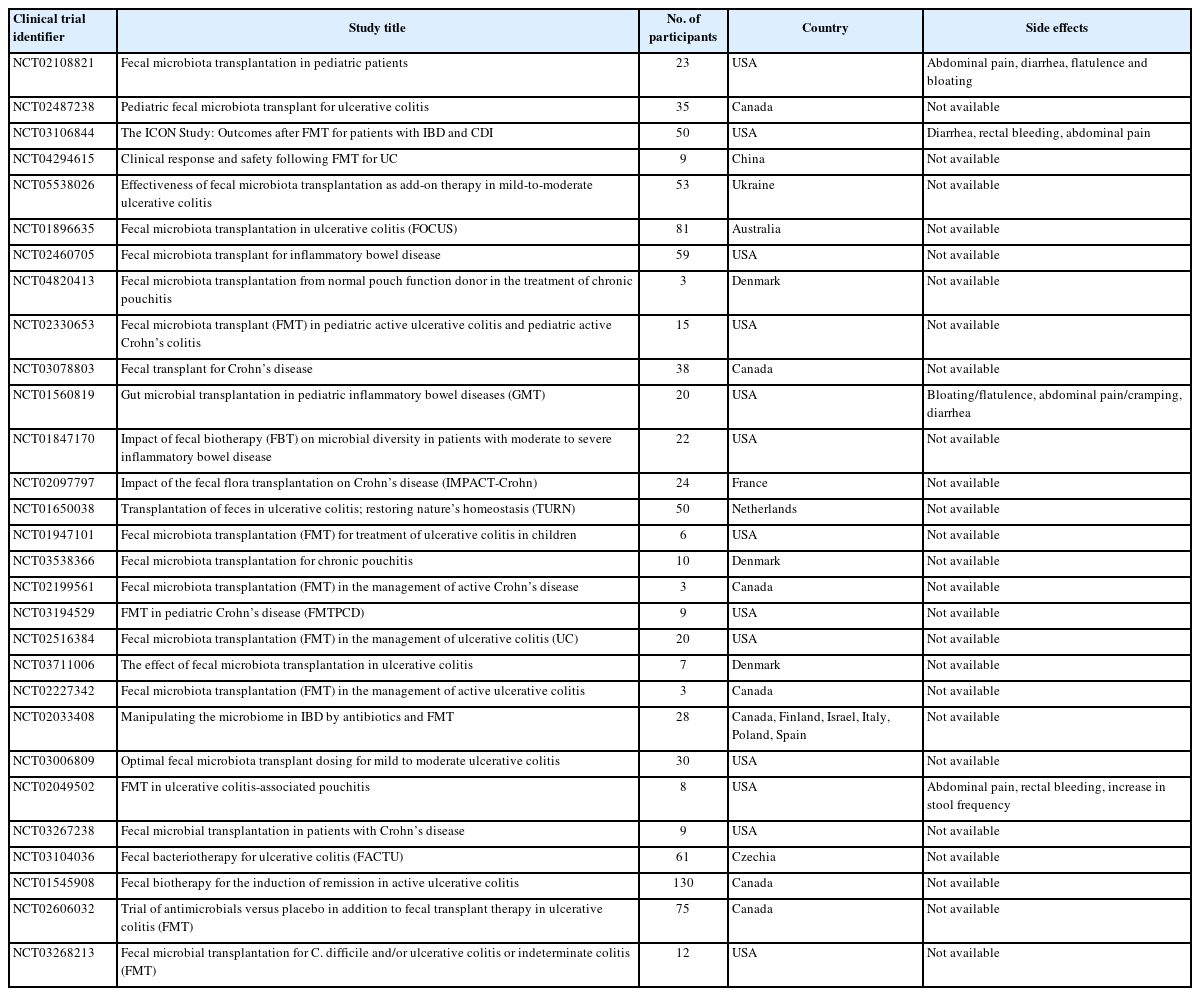
Although a complete cure for IBD is unknown, the current treatment regimen is adopted to reduce inflammation, promote clinical remission, and prevent disease relapse [271-273]. The human gut microbiota is being recognized as a potential therapeutic solution for IBD [274]. Studies have demonstrated that probiotics, prebiotics, and synbiotics effectively modulate the gut microbiota [275-277]. Though promising results have been obtained, much research is needed to establish the efficacy of these treatment options. Clinical trials have shed some light on manipulating gut microbiota through probiotics, prebiotics, synbiotics, and fecal microbiota transplantation (FMT) for IBD treatment (Tables 2 and 3).

Completed Clinical Trials on the Effects of Probiotics, Prebiotics and Synbiotics on Inflammatory Bowel Disease
1. Probiotics
Probiotics are defined as live microorganisms that provide multiple health benefits when administered in adequate amounts [278]. Probiotics deliver many health benefits, modify the gut microflora composition, enhance the intestinal mucosal barrier function, prevent the colonization of pathogenic microbes, and modulate the local and systemic immune responses [279-285]. Accumulating evidence has also revealed that probiotics influence gut microbiota. Since IBD has been linked to dysbiosis of the gut microflora, restoring the normal gut microflora through probiotics is one of the recommended therapeutic options for IBD. Probiotics may prevent IBD by reducing intestinal inflammation [286], downregulating inflammation pathways [287], and producing SCFAs [212]. Studies have found that probiotic therapy can suppress the NF-κB signaling pathway [288,289] and decrease inflammatory cytokines [290].
The most important probiotics for IBD treatment are Bifidobacterium, Lactobacillus, and yeasts [291]. Other probiotics, such as Clostridium butyricum have also been shown to be effective in suppressing inflammation in experimental colitis and are therefore considered for IBD treatment [292]. Although no gold standard regarding effective probiotic doses exists, most commercially available probiotics contain one to 10 billion CFU per dose [290,293]. S. boulardii, Escherichia coli Nissle 1917, and B. breve strain Yakult have shown effectiveness similar to 5-aminosalicylic acid (mesalamine) in maintaining clinical remission in UC patients [294-296]. European Crohn’s and Colitis Organization guidelines mention E. coli Nissle 1917 as an effective alternative to mesalamine in the maintenance of remission in UC patients [297]. Lactobacillus reuteri ATCC 55730, when combined with mesalamine, showed a better clinical response and remission rate in children with UC [298]. Jin et al. [299] found that Lactobacillus plantarum could restore gut barrier function and reduce intestinal inflammation in a mouse model of DSS-induced colitis.
Various probiotic cocktails have been used for IBD treatment. The probiotic VSL#3 is a mixture of 8 bacterial strains: Lactobacillus acidophilus, L. plantarum, Lactobacillus casei, Lactobacillus delbrueckii subspecies bulgaricus, B. breve, Bifidobacterium longum, Bifidobacterium infantis, and Streptococcus salivarius subspecies thermophiles [300]. Studies have found the effectiveness of VSL#3 in inducing remission in patients with mild-to-moderately active UC [301,302]. VSL#3 can also reduce inflammation in pouchitis [303,304]. Studies on a mouse model have shown that VSL#3 effectively reduces ileitis [305] and 2,4,6-trinitrobenzenesulfonic acid-induced colitis [306]. Fedorak et al. [290] reported that VSL#3 could reduce endoscopic recurrence after surgery for CD. Miele et al. [307] showed that combining VSL#3 with 5-aminosalicylic acid (mesalamine) and steroids could significantly improve the remission rate in children with UC. Huynh et al. [308] also showed that combining VSL#3 with standard therapy could improve histological scores in children with UC. A meta-analysis by Mardini and Grigorian [309] suggested that VSL#3 in combination with standard therapy was more effective than standard therapy for UC.
Administration of a cocktail of L. acidophilus, L. plantarum, Bifidobacterium lactis, and B. breve has been shown to enhance the production of intestinal mucus and goblet cells in mice [310]. Another cocktail mixture of L. plantarum, L. acidophilus, Lactobacillus rhamnosus, and E. faecium can increase wound healing and enhance the integrity of tight junctions of epithelial cells [311]. Chen et al. [312] reported that the probiotic mixture of B. infantis, L. acidophilus, and Enterococcus faecalis with or without Bacillus cereus could restore the relative abundance of Lactobacillus, Bifidobacterium, Bacteroides, and Akkermansia in a mouse model of DSS-induced chronic colitis. In UC patients, a mixture of Lactobacillus and Bifidobacterium significantly increases the population of Proteobacteria and reduces Gramnegative rods [313].
When combined with mesalazine S. boulardii, a yeast, can reduce the recurrence rates in CD patients [314]. S. boulardii can also reduce intestinal permeability, increase plasma levels of the anti-inflammatory cytokine, IL-10 and increase intestinal IgA secretion [315]. S. boulardii has also been shown to prevent relapses in patients with CD [316]. Contrary to these studies, Bourreille et al. [317] reported that S. boulardii did not demonstrate any significant efficacy in IBD compared to control individuals.
Next-generation probiotics (NGPs) include human gut commensals that produce some beneficial metabolites [291]. Currently, their use is limited by difficulties in isolation, characterization, cultivation, and formulation. One of the potential NGPs is F. prausnitzii, which has anti-inflammatory properties [318,319]. Several animal experiments have confirmed the therapeutic potential of F. prausnitzii in UC.318,320-322 C. butyricum MIYAIRI, a butyrate-producing bacterium, is effective in preventing pouchitis and alterations of the microbiota in UC patients [83]. Recently, Ma et al. [323] found that C. butyricum MIYAIRI-II could alleviate colitis-related parameters in a mouse model of DSS-induced colitis. NGPs also include genetically engineered probiotic strains. Recently, Zhou et al. [324] genetically engineered E. coli Nissle 1917 to overexpress catalase and superoxide dismutase for treating intestinal inflammation. They found that genetically engineered E. coli Nissle 1917 could effectively alleviate inflammation and repair epithelial barriers in the colon of a mouse model of IBD. They also found that genetically engineered E. coli Nissle 1917 improved the abundance of microbes that maintain intestinal homeostasis, such as Lachnospiraceae and Odoribacter. Many non-conventional probiotics, including Akkermansia muciniphila [325,326], Companilactobacillus crustorum [327], Pediococcus pentosaceus [328,329], Lactiplantibacillus plantarum [330], B. thetaiotaomicron [331], and Christensenella minuta [332], have shown promise in animal studies. More extensive clinical cohort studies are required to evaluate their efficacy in humans.
However, Cochran systematic reviews have concluded that there is no evidence to suggest that probiotics are beneficial for the induction or maintenance of remission in CD [333,334]. Zhang et al. [335], in a systematic review, also concluded that there was no evidence to support the use of probiotics in CD. Studies have also shown that L. rhamnosus GG has no additional benefit over placebo in treating CD [336-338]. Marteau et al. [339] reported that Lactobacillus johnsonii and E. coli Nissle had no significant benefit in maintaining IBD remission. Mallon et al. [340] found that combining S. boulardii and VSL#3 did not significantly improve the remission rates of UC. Matthes et al. [341] reported that the rectal administration of E. coli Nissle for UC had no additional benefits over placebo. Petersen et al. [342] reported that using E. coli Nissle in probiotic therapy had a lower rate of clinical remission in UC. In a randomized controlled trial, Matsuoka et al. [343] reported that probiotics were not effective in maintaining remission in UC.
2. Prebiotics
Prebiotics are food components that are selectively fermented by the gut microflora and help maintain healthy gut microbiota by stimulating the growth of beneficial bacteria [344]. Prebiotics mostly include fructooligosaccharides (FOS), galactooligosaccharides (GOS), and other oligosaccharides, such as pectin [277]. Prebiotics have shown some promise in IBD therapeutics, but some studies have shown contrary results. Prebiotics, such as inulin, have been shown to induce the growth of SCFA-producing bacteria, including Lactobacillus, F. prausnitzii, and Bifidobacterium [345-348]. Inulin has also been shown to improve histological lesions in patients with pouchitis [349]. FOS and GOS can improve the levels of F. prausnitzii [350]. FOS-enriched inulin supplementation can elevate the levels of butyrate [351,352]. FOS are known to increase the population of endogenous microflora, particularly Lactobacillus and Bifidobacterium [353]. FOS are fermented by the gut microbes to produce SCFAs, which exert anti-inflammatory effects [68,354]. Contrary to this, Benjamin et al. [355] reported that the use of FOS in patients with active CD did not improve the remission rate. Casellas et al. [356] found that oligofructose-enriched inulin did not influence the remission rate in patients with UC but could reduce fecal calprotectin levels. The use of prebiotics in IBD treatment is limited by a few studies that are not sufficiently conclusive. More elaborate studies are required to show the efficacy and therapeutic effects of prebiotics for IBD treatment.
Preparations containing both prebiotics and probiotics are called synbiotics [357,358]. Clinical studies [275,357,359,360] and animal studies [361-365] have suggested that synbiotics are effective therapeutic options for IBD. However, contrary results have been obtained in other clinical intervention studies with IBD patients [366-368], raising doubts about their effectiveness.
3. Fecal Microbiota Transplantation
FMT is an emerging biotherapeutic procedure that aims to restore gut microbial ecology by transplanting intestinal microbiota from healthy donors to ameliorate various gastrointestinal disorders [369,370]. FMT enhances the production of SCFAs and restores immune dysregulation [371-373]. FMT has proven successful in treating recurrent Clostridioides difficile infections resistant to antibiotic treatment [374-377].
FMT has gathered much attention recently as a new therapeutic option for IBD (Table 3). Studies have found that FMT is effective in inducing remission in UC patients [373,378-381]. Tian et al. [382] found that after FMT, there was a significant enrichment of Bacteroides, Proteus, and Prevotella and a decline in Klebsiella and Streptococcus. A meta-analysis by Colman and Rubin [383] showed a remission rate of 36.2% in IBD patients who received FMT. They also showed a higher remission rate in CD patients than in UC patients. In a randomized controlled trial by Moayyedi et al. [378], FMT induced clinical remission in patients with active UC. In another randomized placebo-controlled trial for UC by Paramsothy et al. [379], the remission rate of multidonorintensive FMT was higher than placebo. Similar results were obtained by Costello et al. [373] Contrary to these findings, a randomized controlled trial by Rossen et al. [384] failed to find any significant effect of FMT in patients with UC. In a pilot study, Sood et al. [381] concluded that FMT may be one of the therapeutic options for clinical remission in UC patients. Sokol et al. [385] carried out a pilot randomized controlled study to find the effects of FMT in CD patients showing clinical remission with systemic corticosteroids. In this study, the beneficial effects of FMT were evaluated using several clinically relevant endpoints, such as C-reactive protein level and CD Endoscopic Index of Severity. Kunde et al. [386] found significant improvement in 9 children with UC who received FMT via enema. Cui et al. [387] showed that FMT improved clinical outcomes in 57% of patients with steroid-dependent UC. A meta-analysis on FMT for IBD by Paramsothy et al. [372] showed a clinical remission rate of 50.5%. A meta-analysis by Caldeira et al. [388] reported that FMT had a complete remission rate of 37% for IBD patients.
However, various clinical studies conducted to examine the effect of FMT on IBD have obtained inconsistent results, raising doubts about its effectiveness. Available data suggest that the efficacy of FMT in treating IBD is not predictable [389]. Angelberger et al. [390] reported that only one UC patient showed some improvement after 12 weeks of FMT. Similarly, Suskind et al. [391] failed to find any significant improvement in 4 children who received a single FMT through a nasogastric tube. Such discrepant results in clinical trials may reflect the heterogeneity in disease biology, donor selection, FMT regimen, and individual response to the treatment.
The safety and questionable efficacy of FMT have precluded its wider use for IBD treatment. FMT is also associated with certain risks, further limiting its use as a therapeutic option [16]. The success rate of FMT in IBD treatment can be enhanced by standardizing various parameters, such as donor selection, route and mode of administration, and optimal dose and frequency of infusions.
CONCLUSIONS AND FUTURE PERSPECTIVES
The human gut is a complex ecosystem with a continuous crosstalk between microbes and host cells. A growing body of evidence has indicated a close association between alterations in gut microbiota and the pathophysiology of IBD. Furthermore, the microbial metabolites may also affect the progression of IBD. Since different studies provide inconsistent, even contradictory results, further studies must be carried out to reach a consensus. Advances in next-generation sequencing technology have enabled deep insights into the complex microbial communities inhabiting the human gut and have helped identify and compare the normal gut microbiota with the altered microbiota in IBD. Also, metabolomics has helped identify altered microbial metabolites in IBD patients. Further research describing the role of the gut microbiota in maintaining gut barrier function and the immune system is essential to understand IBD pathophysiology. The findings from such studies must be translated into clinical practice.
There is no known cure for IBD. The current treatment options help induce clinical remission by reducing inflammation. Though still in its infancy, targeted microbiota manipulation using probiotics, prebiotics, synbiotics, and FMT is one of the potential armamentariums in IBD. Although promising results have been shown for many probiotics, including VSL#3 and E. coli Nissle 1917, clinical trials with larger cohorts must be carried out to reach conclusive evidence. NGPs, including engineered microbes, may provide a new direction for IBD treatment. Another microbiota-based intervention, FMT, has gathered much attention as a new therapeutic option for IBD, but its efficacy remains questionable. Standardization of various parameters will surely help increase its effectiveness and success rate in IBD. Future research should be directed towards utilizing gut microbiota for developing less invasive diagnostic tools for IBD and microbiota replacement as a potential therapeutic for IBD treatment.
Notes
Funding Source
The authors would like to acknowledge the NTU Start Up Grant (021337–00001), Wang Lee Wah Memorial Fund, and Singapore Ministry of Education (MOE) Tier 1 Academic Research Fund (RG37/22) for support of this work.
Conflict of Interest
No potential conflict of interest relevant to this article was reported.
Data Availability Statement
Not applicable.
Author Contribution
Conceptualization: Pandey H, Wong SH, Lal D. Resources: Pandey H, Jain D, Tang DWT, Wong SH, Lal D. Diagrams: Tang DWT. Supervision: Lal D, Wong SH. Writing-original draft: Pandey H, Jain D, Tang DWT, Wong SH, Lal D. Writing-review & editing: Pandey H, Jain D, Tang DWT, Wong SH, Lal D. Approval of final manuscript: all authors.